Venomous animals, in all their glorious shapes and sizes, have been the abiding passion of my life. Venom systems are key evolutionary innovations that have arisen independently in a myriad of animal groups. The most extensively studied venoms are from the medically-important snakes, yet venom systems are also present in a diverse range of other animals including lizards (Gila Monster, Komodo Dragon), mammals (platypus, shrews), fish (stingrays, stonefish), molluscs (octopus, cone snails), cnidarians (blue-bottle, box jellyfish) and echinoderms (Crown of Thorns starfish, flower urchin). Venom systems have also evolved independently within several arthropod groups such as centipedes, scorpions, spiders, stabbing insects and stinging insects.
I am working on a general theory of how venom evolves by applying all the theories I developed on reptile venom evolution, to other orders. I am currently concentrating on the molecular evolution of venom proteins in cephalopods (cuttlefish, nautiluses, octopuses and squids) from tropical and temperate waters. As with snakes and lizards, there are many more venomous cephalopod species than appreciated.
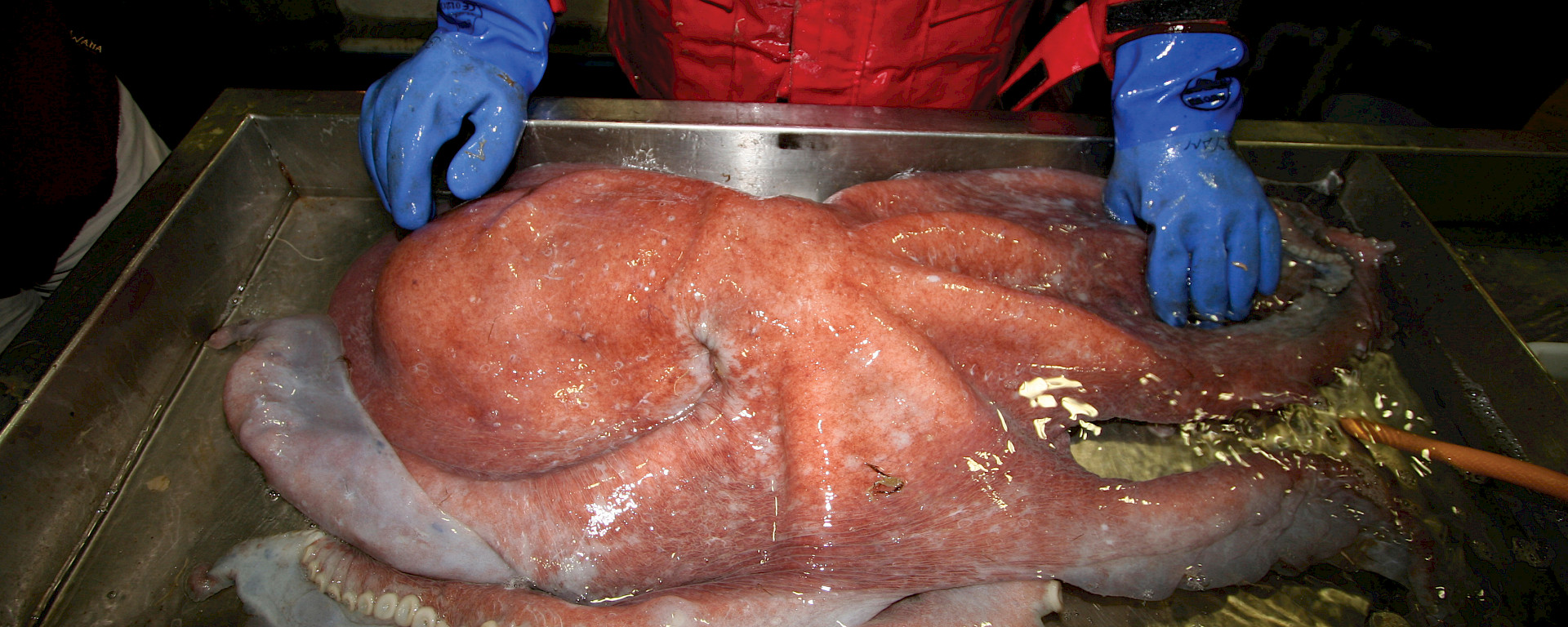
During the recent Collaborative East Antarctic Marine Census I was astounded at the biodiversity of the Antarctic waters. The tropics are always portrayed in documentaries as marine hotspots. They are, of course, much more accessible and amenable for research than Antarctic waters. But the sheer quantity and diversity of life in the Southern Ocean rivals anything I have seen on my expeditions to the Great Barrier Reef, Coral Sea, Asia, the Caribbean, or other locations in-between. Two venomous lineages — octopuses and anemones — were very well represented, both in total numbers and species diversity.
After our successful expedition we now have a long, hard slog in the laboratory ahead of us. The first step is to DNA bar code (genetically fingerprint) each octopus in order to reveal the biodiversity present in the Antarctic waters. We have already completed this aspect and are currently analysing the results.
The second step will be to obtain a snapshot of every protein being produced at the time the venom glands were removed. I have already achieved this with octopuses, cuttlefish, centipedes, fish, snakes and even the venom glands of the iconic Komodo Dragon. My studies of octopus and cuttlefish venom glands show that they have toxic proteins in common, resulting from early ‘recruitment’ events into the chemical arsenal of their common ancestor. However, during the subsequent evolution of the animals themselves, they have recruited new proteins into the chemical arsenal that are unique to each lineage. This shows that while cephalopods share a common venomous ancestor, the venom system continues to diversify.
The third step in the process will identify the spectrum or range of proteins in the venom and their relative quantities. The glands of each specimen will be analysed separately to reveal their unique molecular fingerprints, thus revealing any regional variation within and between species.
These results will guide bioactivity testing to determine the effects of the crude venom and purified individual toxins on living cells. The activity of the crude venom is greater than the sum of the individual components, due to the synergistic action that often occurs between different molecules.
Of particular interest will be the gene sequences and actions of enzymes from cold water cephalopod species (from Antarctica and Norway), with the same types of enzymes from temperate and tropical species. Temperature is a major variable in the action of enzymes and most enzymes have a narrow ‘sweet spot’ of optimal activity. As the same enzyme classes are shared between tropical and Antarctic species, significant biochemical changes must exist between them in order for activity to occur at the radically different temperatures (32ºC versus –2ºC). The fundamental changes responsible for the activity at different temperatures will be elucidated by comparing differences in the sequence of amino acids (protein building blocks) and then making chimeric versions of the proteins in the laboratory to determine which amino acids confer the temperature-dependent activities.
In our studies of temperate and tropical species so far, we have discovered new small proteins with very intriguing activities, which are potentially useful in drug design and development. We will be searching for new small proteins in the venoms of the specimens we have collected on this expedition.
The trip to Antarctica was not only a professional bonanza but also as close to a religious experience as I will ever get. I hope one day to return to the mecca of the frozen South.
*In addition to the award of berths by the Australian Antarctic Division, this research is funded by the Australian Research Council, Australian Academy of Sciences (J.G. Russell Award), the CASS Foundation and the Australian & Pacific Science Foundation.
BRYAN GRIEG FRY
Group Leader, Venomics Research Laboratory,
Department of Biochemistry & Molecular Biology,
Bio21 Institute, University of Melbourne
After completing a PhD on the biochemistry of snake venoms at the University of Queensland’s Drug Design and Development Centre, Dr Bryan Fry undertook a two year postdoc at the National University of Singapore. He then joined the Australian Venom Research Unit as Deputy Director (Research) for four years. He now leads his own venom research group within the University of Melbourne’s Department of Biochemistry and Molecular Biology. Bryan spends a great deal of time traveling the world to collect and milk venomous snakes and other animals. His work has lead to the discovery that the number of venomous snakes in the world is around 2700, not 250, as was originally accepted. He has also found that the evolution of reptile venom goes back much further than previously thought — approximately 200 million years– and that the toxic effects caused by the bites of the Komodo Dragon are caused by venom, not by toxic bacteria.